Super-chilled imaging technique brings its developers the Nobel Prize in chemistry
It allows scientists to now map biological molecules, such as proteins, clearly and on the atomic scale
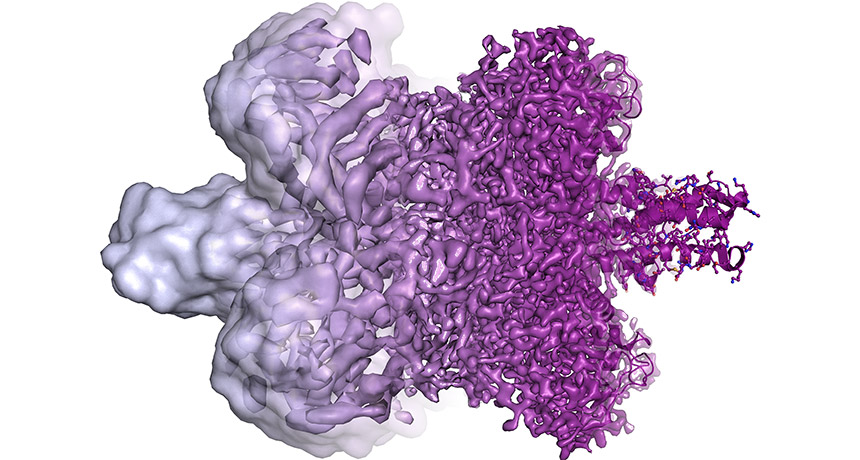
Cryo-electron microscopy is an imaging technique that involves flash freezing molecules to see their structures. This technique has just won its inventors the Nobel Prize in chemistry. The technique has dramatically improved over time. Molecules that once appeared as blobs (left) now appear in all their complex glory (right).
© MARTIN HÖGBOM/STOCKHOLM UNIV.
Share this:
- Share via email (Opens in new window) Email
- Click to share on Facebook (Opens in new window) Facebook
- Click to share on X (Opens in new window) X
- Click to share on Pinterest (Opens in new window) Pinterest
- Click to share on Reddit (Opens in new window) Reddit
- Share to Google Classroom (Opens in new window) Google Classroom
- Click to print (Opens in new window) Print
By Carolyn Gramling and Laurel Hamers
Three developers of a technique to view proteins, viruses and other biological objects have just won the 2017 Nobel Prize in chemistry. The technique requires flash freezing the materials. Now, with some recent advances, scientists can use this technique to peer into structures at the atomic scale.
Jacques Dubochet works at the University of Lausanne in Switzerland. Joachim Frank is a scientist at Columbia University in New York City. Richard Henderson works in England at the MRC Laboratory of Molecular Biology in Cambridge. Together, they contributed to the development of what’s now known as cryo-electron microscopy.
The Royal Swedish Academy of Sciences announced the new winners on October 4. At an awards ceremony in December, the men will receive a medal. They’ll also share a 9-million-Swedish-kronor prize. (It’s worth about $1.1 million.)
Andrew Murray is a systems biologist at Harvard University. He applauds the selection of these winners. Their work “lies at the basis of an incredibly important technique.” The award, he says, goes to people who helped science be able to “see molecules up close and personal.”
Now we can see the intricate details of every drop of our body fluids, Sara Snogerup Linse said at a news conference today. With that, “we can understand how they are built and how they act and how they work together,” said Linse. She chaired the 2017 chemistry Nobel committee. “Soon,” she predicted, there will be “no more secrets.”
Story continues below image.

Evolution of the technique
The technique allows scientists to map the landscape of molecules. They can see details at scales as tiny as just tenths of a nanometer (billionths of a meter). Such closeup details can help them see how these very small things work.
In the past, structural biologists have relied on two other tools. One of them is nuclear magnetic resonance (NMR) spectroscopy. NMR allows scientists to probe biological molecules in a solution, such as water. But this technique generally could be used only with fairly small proteins. The other is X-ray crystallography. It bounces X-rays off of a crystal to create a 3-D picture of a molecule’s structure. Unfortunately, “A lot of proteins cannot be crystallized,” Murray notes, “So their structures are inaccessible to the prying mind of the biochemist.”
Another technique is electron microscopy (EM). It works well for imaging nonliving things. It fell short, however, when applied to biological molecules.
EM is similar to X-ray crystallography in some ways. The technique maps how the beams deflect off of the material they’ve passed through to sketch out the material’s atomic structure. But the powerful electron beams can incinerate any protein or biological material as they pass through it. This produces fuzzy images. And there’s another drawback. The material being imaged must be in a vacuum (in the microscope). That causes water within biological molecules to evaporate. This further distorts the shapes.
In 1975, to get around these problems Henderson placed bacterial proteins within the electron microscope. To keep them from drying out, he covered their membrane with a sugar-water solution. To minimize damage to the proteins, he also weakened the electron beam. Still, the images were fuzzy, not clear.
But the proteins in his sample happened to be oriented in the same direction. So Henderson put the fuzzy images together. This created one sharper image.
He then turned the membrane this way and that. With each change in direction, he grabbed new images. Depending on how he added them together, he could now see the protein’s structure in three dimensions. This was better than before. But it still didn’t give him atomic-scale details.
Frank’s work helped with that. He was tackling the issue of orientation. Unlike Henderson’s proteins, most molecules do not all line up in the same direction. Frank’s solution, published in 1981, was to create a computer algorithm. He scanned the images to look for any proteins oriented in the same direction. Then the computer program grouped similar ones together. Like Henderson’s combined image, each group that Frank processed was somewhat sharper than before.
Meanwhile, Dubochet was probing how to keep his samples from drying out and becoming damaged. Freezing the samples also wouldn’t work. That’s because ice crystals will distort a molecule’s shape. But if the water cooled superfast to ‒196° Celsius (‒320° Fahrenheit), it would turn solid without forming ice crystals. Instead, it became glass-like, or vitrified. In 1984, Dubochet showed that when he covered viruses with a fine layer of vitrified water, he could be safely image them in an electron microscope.
This was the birth of cryo-electron microscopy, or cryo-EM.
At last, atomic-scale resolution
In 1990, Henderson produced the first cryo-EM image of a protein at the atomic scale. The next year Frank used cryo-EM to view ribosomes (RY-boh-soams). These are particles that help make proteins. But for some time, such images were not all that sharp. Many researchers in fact took to calling this technique “blobology.”
Over time, however, other researchers offered up improvements. Some used better optics or detectors or computing techniques. Sriram Subramaniam is a structural biologist at the National Cancer Institute in Bethesda, Md., In 2015, he reported improving the resolution of cryo-EM images to an ultrafine 0.22-nanometer scale. This rivalled the resolution of the current gold standard: X-ray crystallography.
“We are now at atomic resolution,” says Subramaniam. “Now, we need to apply this to look at larger [structures] to understand how these different molecular machines work.”
Recently, cryo-EM has begun to demonstrate its value. Last year, for instance, it was used to map the structure of the Zika virus. This helped identify possible regions of the virus that could be targeted with a vaccine or other drug.
New types of electron detectors have recently been developed for this type of microscope, notes Michael Rossmann. He’s a physicist and microbiologist at Purdue University in West Lafayette, Ind. He worked on the Zika mapping. With these new advances, he says, “Cryo-EM has revolutionized structural biology, particularly in the last three years.” He describes it as a “resolution revolution.” Next up, he predicts: looking at an entire cell.
Subramaniam also believes there are exciting years ahead. “This is just the beginning.”






